|
LBIBCell
|
|
LBIBCell
|
LBIBCell (Lattice Boltzmann Immersed Boundary) is a 2D simulation software to study the interplay between cell and tissue mechanics and biomolecular signaling. Here, you will find information how to download and build the software, extensive tutorials, and the detailed documentation of the source code. For more information about the implemented algorithms and applications, please turn to the corresponding publications.
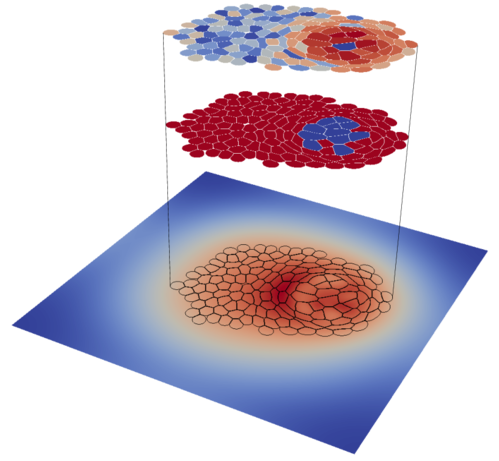
We highly recommend to go through the tutorial examples in order to get to know how to set up your own scenarios and implement custom modules.
The following graph shows how the tutorials depend on each other:
You can download the softwawre from here.
Use cmake and make. For more details see Installation.
For more details see Dependencies
The software is free and open source. It is distributed under a MIT license (see The licence for the library)
Tanaka S, Sichau D, Iber D, LBIBCell: A Cell-Based Simulation Environment for Morphogenetic Problems. Bioinformatics, in press.
Please check out our group website.